Cancer researchers identify protein with novel anti-tumoral activities
Understanding how cancer develops is critical for designing effective, personalized cancer therapies. Researchers have known for years that cancer begins with mutations in certain types of genes. One of these types of cancer genes are so-called “tumor suppressors.” When functioning normally, tumor suppressor genes can stop malignant cells from undergoing uncontrolled cell proliferation and initiate a process of cell elimination called apoptosis, a form of cell death.
Mutations in tumor suppressor genes can cause these genes to lose their functionality, eventually contributing to the development of cancer.
In a recent study published in Cell Reports, researchers at the University of Colorado Anschutz Medical Campus described the discovery and characterization of a novel protein involved in a mechanism that suppresses different types of tumors.
The tumor suppressor gene called TP53 effectively restricts the development and growth of many different tumor types across the human body, and it is the most frequently mutated tumor suppressor gene in human cancers. This gene encodes a protein called p53, which is both a potent inhibitor of cell proliferation and an inducer of apoptosis.
According to Zdenek Andrysik, Ph.D., assistant research professor of pharmacology in the University of Colorado School of Medicine and one of the authors in the paper, “In more than half of cancer cases, TP53 is not mutated, remaining instead in a dormant state. Accordingly, many research efforts have been devoted to the development of drugs that could reactivate this latent form of p53 for cancer therapy.”
“However, most cancers respond to activation of p53 with these medications with a transient block in cell proliferation. A better response to these drugs would be cancer cell elimination via apoptosis. Therefore, it is critical for us to understand what other factors are required for an effective p53-targeted cancer therapy.”
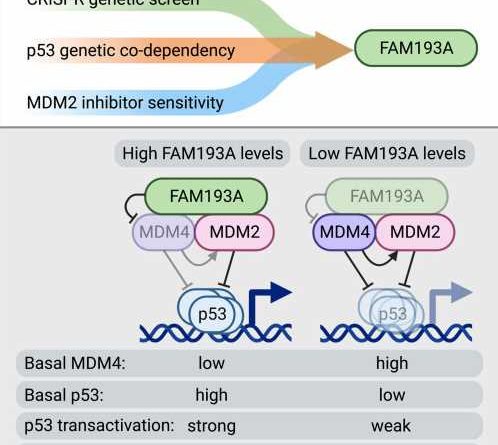
To address this knowledge gap, Maria Szwarc, Ph.D., and Anna Guarnieri, Ph.D., former postdoctoral fellows in the department of pharmacology and co-leading authors of the paper, employed a multi-disciplinary experimental approach, including genetic screening using CRISPR technology, to disrupt all genes in the human genome one-by-one and pinpoint which genes are required for full p53 activation.
As a result, the screening identified the protein called FAM193A, about which very little was known, as a potent and widespread positive regulator of p53 activity.
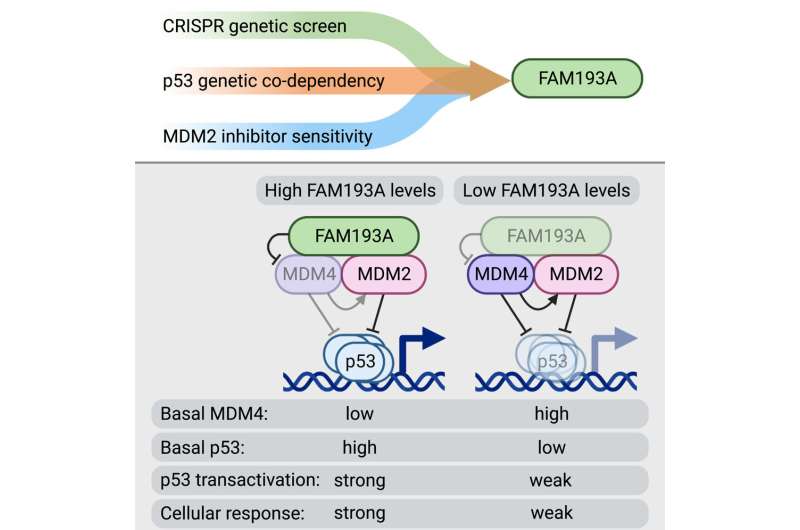
“Our follow-up studies revealed that FAM193A is required for the stabilization of the p53 protein and its functionality,” Dr. Szwarc explains.
“The results showed that FAM193A interferes with cellular factors that usually repress p53 function, the proteins MDM2 and MDM4, which are commonly overactive in cancers. We found that FAM193A can antagonize the MDM4 protein and thus unleash p53’s ability to stop cancer cell proliferation and survival. In agreement with these findings, we documented that high levels of FAM193A found in certain tumor types is associated with a better prognosis for cancer patients.”
Joaquin Espinosa, Ph.D., a professor of pharmacology in the University of Colorado School of Medicine, director of the Linda Crnic Institute for Down Syndrome, and head of the research team states, “There was very little, if any, information about FAM193A function when we identified it in our genetic screen as a partner of p53.”
“However, advanced computational analyses of large-scale datasets across hundreds of cancer cell types and human tumors revealed a clear functional connection between p53 and FAM193A. These discoveries bring us one step closer to effective p53-based cancer therapies, which should consider FAM193A. Future research will be aimed at finding ways to bolster the activity of these partner factors for effective tumor suppression.”
More information:
Maria M. Szwarc et al, FAM193A is a positive regulator of p53 activity, Cell Reports (2023). DOI: 10.1016/j.celrep.2023.112230
Journal information:
Cell Reports
Source: Read Full Article