CASdetec: new CRISPR-based SARS-CoV-2 detection platform detects nucleic acids with high sensitivity and specificity
Coronavirus disease 2019 (COVID-19), caused by the novel coronavirus named severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), has caused a global pandemic of unprecedented proportions, overwhelming public health systems and economies across the world.
Detection of viral nucleic acid has been an important strategy in the early diagnosis of SARS-CoV-2. Four CRISPR-based protocols have been reported so far for detecting SARS-CoV-2. RNA samples with over 1×104 to 1×105 copies/mL (SHERLOCK) or 1×104 copies/mL (DETECTR) can be detected in less than an hour with the help of lateral flow protocols.
New CRISPR-assisted nucleic-acid detection platform – CASdetec
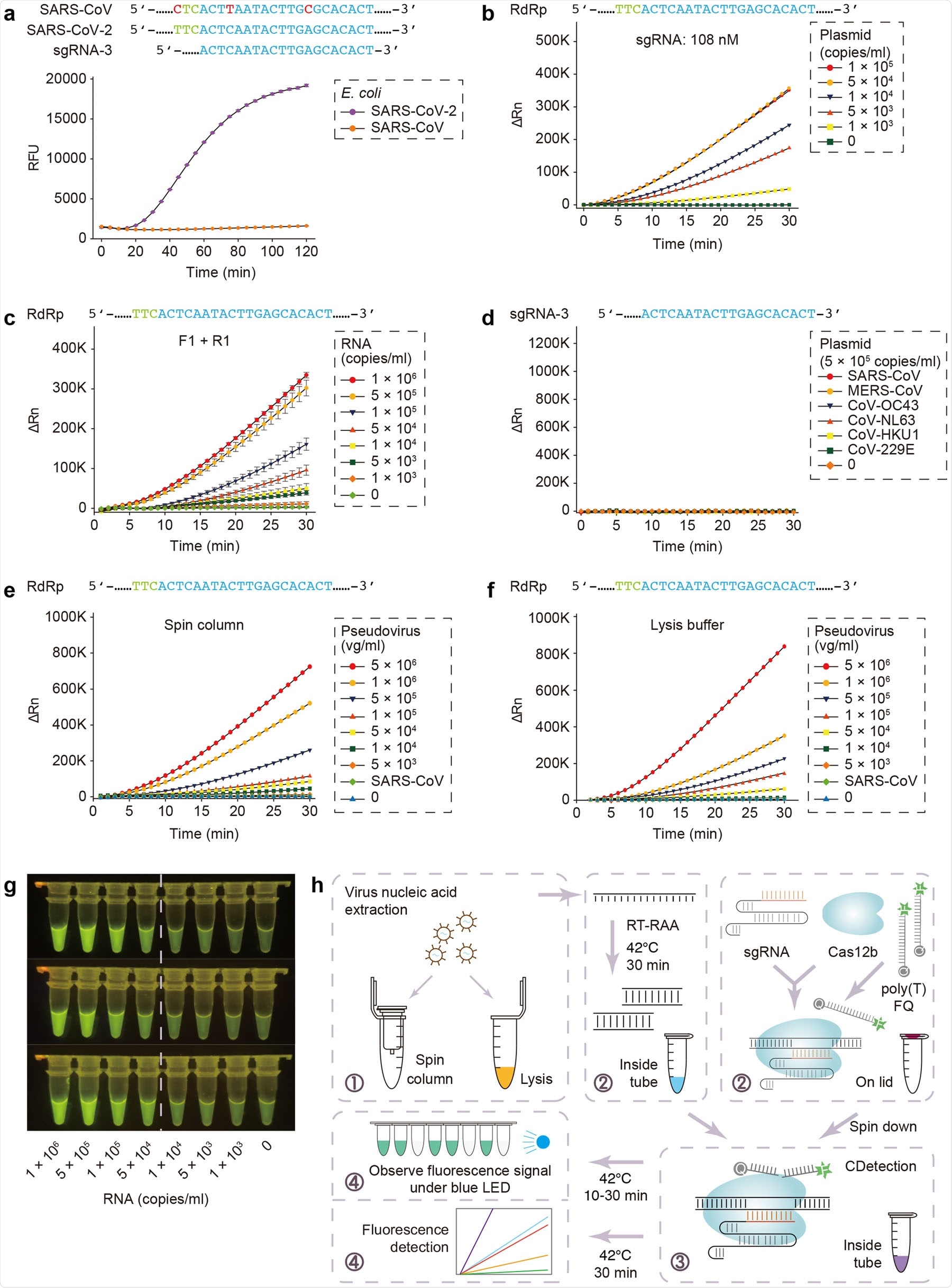
To add to existing CRISPR-based nucleic acid detection protocols, a team of researchers recently established a new SARS-CoV-2 detection protocol based on their previously reported platform – CDetection (Cas12b-mediated DNA detection). They combined nucleic-acid amplification methods and sample treatment protocols with CDetection and established an integrated nucleic acid detection platform called CRISPR-assisted detection or CASdetec. CASdetec has a detection limit of 1×104 copies/mL for SARS-CoV-2 pseudovirus, without any observed cross-reactivity.
According to the available SARS-CoV-2 genome sequence, they designed 7 sgRNAs around the RdRp locus, as per World Health Organization (WHO) recommendation. Due to the high similarity of SARS-Cov-2 to SARS-CoV, they conducted initial experiments on the plasmids of both viruses. Fluorescence kinetics studies show that sgRNA-3 was distinct in that it could distinguish between the 2 coronaviruses and also produced the most distinct fluorescence signal.
The chosen sgRNA and primers can be used for most of the reported SARS-CoV-2 genomes
Since CRISPR cannot detect DNA at <1–10 nM of amplification product in the reaction mix, increasing molecular collisions between the target and CRISPR is crucial to improve the test's sensitivity. The researchers found that increasing the concentration of sgRNA by 3 folds enhances the fluorescence signal as well as the signal-to-background ratio. Moreover, it also boosts the rate of reaction. They designed and screened RAA primers to match their previously optimized sgRNA-3. The screenings showed that by using the best primer pairs with sgRNA-3, detection of nucleic acid is possible in SARS-CoV-2 RNA samples with 5 × 103 copies/mL.
The researchers also analyzed all 1792 SARS-CoV-2 sequences on GISAID until 26 March 2020 and found that 1673 of those sequences match their chosen primers and sgRNA. Only 3 of them has 1 mismatch to the forward primer, and only 2 of them had 1 mismatch to the reverse primer, which suggests that the sgRNA and primers they have chosen can be used for most of the reported SARS-CoV-2 genomes.
In order to validate the specificity of this new method for detection of SARS-CoV-2 nucleic acid, they tested the protocols against 6 coronaviruses – SARS-CoV, MERS-CoV, CoV-HKU1, CoV-229E, CoV-OC43, and CoV-NL63 – that cause respiratory illnesses. The results show no cross-reactivity with other endemic coronavirus and suggested that their sgRNA and primers had high sensitivity and specificity.
CASdetec assay design and optimization process could offer guidance for future CRISPR-based nucleic acid detection
To summarize, the authors established a new platform for nucleic acid detection based on CRISPR – the CASdetec. The platform has many procedures, including nucleic acid amplification, virus handling, and CRISPR-based detection. CASdetec is capable of detecting pseudovirus samples with >1 × 104 copies/mL without any cross-reactivity to other coronaviruses.
Additionally, the researchers also optimized the workflow to perform both reactions in a single tube without opening the lid. This will help prevent aerosol contamination and decrease the rate of false-positive test results.
According to the authors, in order to ensure the accuracy of CASdetec, users need to be careful to avoid nucleic acid aerosol contamination. They can do the following to avoid contamination: (a) perform sample treatment, reagent preparation, amplification, and product analysis in different rooms; and (b) carefully handle nucleic acid samples and reagents to avoid contamination through touch.
The researchers recently presented their assay design and optimization process in a Correspondence in the journal Nature. They believe that this process could offer guidance for future nucleic acid detection assay development and optimization based on CRISPR.
The team concludes:
We have established a CASdetec platform, which consists of procedures including virus handling, nucleic acid amplification, and CRISPR-based detection."
- Guo, L., Sun, X., Wang, X. et al. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov 6, 34 (2020). https://doi.org/10.1038/s41421-020-0174-y, https://www.nature.com/articles/s41421-020-0174-y
Posted in: Medical Science News | Medical Research News | Disease/Infection News | Healthcare News
Tags: Assay, Contamination, Coronavirus, Coronavirus Disease COVID-19, CRISPR, Diagnostics, DNA, Fluorescence, Genome, Health Systems, Locus, MERS-CoV, Nucleic Acid, Pandemic, Pseudovirus, Public Health, Reagents, Respiratory, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Virus

Written by
Susha Cheriyedath
Susha has a Bachelor of Science (B.Sc.) degree in Chemistry and Master of Science (M.Sc) degree in Biochemistry from the University of Calicut, India. She always had a keen interest in medical and health science. As part of her masters degree, she specialized in Biochemistry, with an emphasis on Microbiology, Physiology, Biotechnology, and Nutrition. In her spare time, she loves to cook up a storm in the kitchen with her super-messy baking experiments.
Source: Read Full Article
