How understanding the dynamics of yeast prions can shed light on neurodegenerative diseases
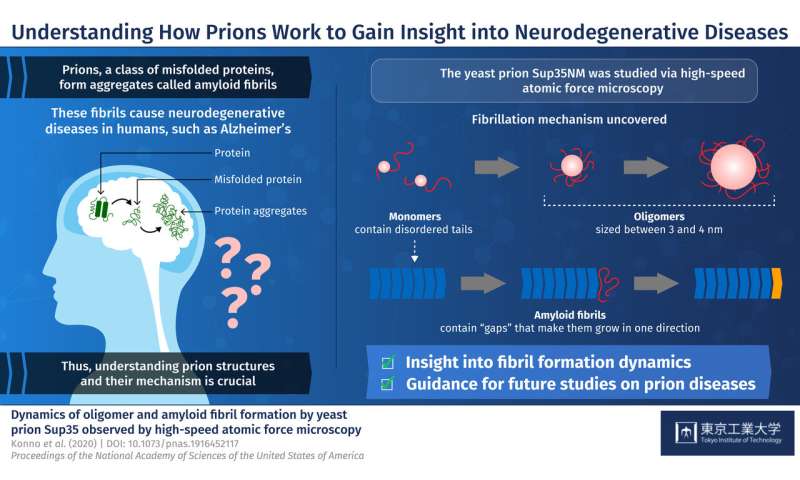
Prions are a class of misfolded proteins that form aggregates called amyloid fibrils. These aggregates are the main culprit behind severe mammalian neurodegenerative diseases like Alzheimer’s. What makes them so deadly is that they are capable of transmitting their erroneous conformation to otherwise healthy proteins, causing an imbalance in cellular function. Currently, there are no effective treatments for fatal prion diseases, mainly because studying mammalian prions is challenging. Thus, scientists have turned to studying prions in less complex organisms like yeast, which could provide more insight about mammalian prions. Yeast prions, such as the protein Sup35NM, are also known to form amyloid fibrils. But the process by which individual Sup35NM molecules, called monomers, combine to form amyloid fibrils is not yet understood. Moreover, Sup35NM monomers sometimes form smaller structures called oligomers—another process that remains unclear. To shed light on prion-related diseases, it is crucial to understand the exact details of how amyloid fibrils are formed from prions.
In a new study published in PNAS, researchers from Tokyo Institute of Technology and Kanazawa University led by Prof Hideki Taguchi set out to dig deeper into prion structures and their mechanisms. They used a modern microscopy technique called high-speed atomic force microscopy (HS-AFM) developed by Dr. Toshio Ando at Kanazawa University to visualize the formation of Sup35NM amyloid fibrils in real time. Prof Taguchi explains, “Previous studies used methods that do not allow for the simultaneous, high-resolution assessment of the structures and dynamics involved in Sup35NM oligomerization and fibrillation. To overcome this limitation, we used HS-AFM, which allows the direct visualization of protein molecules in dynamic action at a high resolution.”
To begin with, the researchers analyzed Sup35NM monomers using HS-AFM. Their analysis revealed that Sup35NM molecules contained a globular structure with two highly flexible tail-like structures. Then, to observe the oligomeric forms of Sup35NM, monomers were incubated in a denaturing solution in controlled conditions, which, in a few hours, led to the formation of oligomers. Further HS-AFM analyses revealed the exact size of these oligomers—a maximum of 3–4 nm, not growing any longer than this. Thus, the scientists inferred that this size could be an inherent property of oligomers under these conditions.
The researchers then moved on to study the fibril forms of Sup35NM. As opposed to oligomers, fibrils required much a longer incubation time to form, typically two to three days. In addition, the researchers observed that the fibril elongated smoothly without the attachment of the oligomers. These findings indicated that Sup35NM oligomers are not a prerequisite for the formation of amyloid fibrils, leading the scientists to conclude that oligomers might actually possess cellular functions that are different from those of Sup35NM.
Finally, the researchers studied an interesting phenomenon where oligomers and fibril structures seem to maintain an interspatial distance or “gap.” They observed that the trunk of the fibrils has a rigid structure that repels oligomers coming from the sides. Meanwhile, the gap between the tip of the fibril and nearby oligomers is less pronounced because the tip does not have such rigid structures. This helped the scientists understand why Sup35NM amyloid fibrils always grow in a straight direction without forming any branches.
In sum, these findings provide a detailed account into the structural and functional characteristics of yeast prions in their monomeric, oligomeric and fibril forms. “Our HS-AFM observations of Sup35NM unveiled features of Sup35NM dynamics, providing mechanistic insight into amyloid fibril formation,” concludes Prof Taguchi.
Source: Read Full Article
