In vitro zoo helps in understanding SARS-CoV-2
Since the beginning of the pandemic, several reports have indicated that SARS-CoV-2 spillover events have occurred from humans to animals, as evidenced by the transmission of the virus between keepers and tigers and lions in the Bronx Zoo in New York. However, to date, the full range of animal species that are susceptible to SARS-CoV-2 infection remains unclear. Typically, such information could be obtained by experimentally infecting a large variety of animal species with SARS-CoV-2 to see if they are susceptible. However, in order to reduce and refine such animal experiments, the researchers at the University of Bern and at the IVI set out to answer this question using a different, and more animal friendly approach.
Building an in vitro zoo
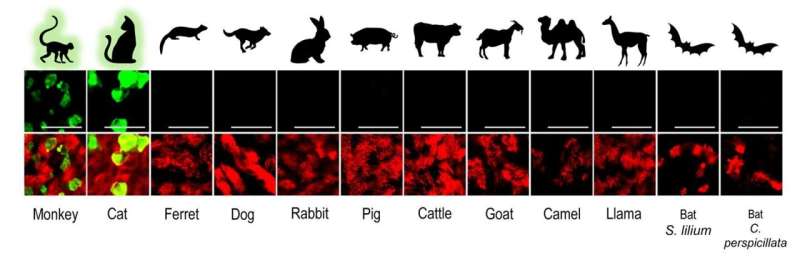
The authors used their knowledge on advanced in vitro cell culture models for the human respiratory tract, to create a large collection of similar models from various domesticated and wildlife animal species. For this the team isolated airway epithelial cells (AEC) from tracheobronchial tissue from deceased animals and created a cell biobank from different animal species that can be used to establish well-differentiated AEC culture models to determine whether these animals can be infected with SARS-CoV-2. Because the cells are isolated from deceased animals and the number of isolated cells can be increased in a petri dish, the researchers do not have to perform any animal experiments. To date, the cell biobank contains primary cells from 12 different animal species; rhesus macaque, cat, ferret, dog, rabbit, pig, cattle, goat, llama, camel, and two neotropical bat species. “Our collection is unique, and thus far we are the first that have used such a large collection of advanced in vitro cell culture models from various domesticated and wildlife animal species to assess their susceptibility to SARS-CoV-2 infection,” says Ronald Dijkman from the Institute for Infectious Diseases (IFIK) at the University of Bern.
Rhesus macaques and cats as potential spillback reservoirs for SARS-CoV-2
In these experiments, the researchers found that the in vitro results agreed well with previously published studies using animal experimentation to assess the susceptibility of different animals to SARS-CoV-2 infection. Using whole viral genome sequencing, the researchers also observed that SARS-CoV-2 replicated in the in vitro models of monkey and cats, without the need for the virus to adapt. These findings suggest that certain species of monkeys and cats may be particularly vulnerable to SARS-CoV-2 infection. “Our findings, together with the reports from previously documented spillover events, indicate that close surveillance of these animals and other close relatives, whether they live in the wild, captivity or households, is necessary,” says Dijkman.
This information can be used by the responsible authorities such as the Swiss Federal Office for Public Health and the Federal Food Safety and Veterinary Office, for SARS-CoV-2 surveillance at the human—animal interface. Specifically, it helps them to establish and tailor early detection surveillance programs to monitor animals that can act as potential spillback reservoirs for SARS- CoV-2. Dijkman added, “This will benefit the general public since it will help prevent new SARS- CoV-2 variants from developing in animal reservoirs and potentially being reintroduced into the human population, to which the current vaccines may not be protective.”
Implementing the 3R (replace, reduce, refine) principles in coronavirus research
Source: Read Full Article
