Machine learning fuels personalized cancer medicine
The Biomedical Genomics laboratory at IRB Barcelona has developed a computational tool that identifies cancer driver mutations for each tumor type.
This and other developments produced by the same lab seek to accelerate cancer research and provide tools to help oncologists choose the best treatment for each patient.
The study has been published in the journal Nature.
Each tumor—each patient—accumulates many mutations, but not all of them are relevant for the development of cancer. Researchers led by ICREA researcher Dr. Núria López-Bigas at IRB Barcelona have developed a tool, based on machine-learning methods, that evaluates the potential contribution of all possible mutations in a gene in a given type of tumor to the development and progression of cancer.
In previous work already available to the scientific and medical community, the laboratory developed a method to identify those genes responsible for the onset, progression, and spread of cancer. “BoostDM goes further: it simulates each possible mutation within each gene for a specific type of cancer and indicates which ones are key in the cancer process. This information helps us to understand how a tumor is caused at the molecular level and it can facilitate medical decisions regarding the most appropriate therapy for a patient,” explains Dr. López-Bigas, head of the Biomedical Genomics lab. In addition, the tool will contribute to a better understanding of the initial processes of tumor development in different tissues.
The new tool has been integrated into the IntOGen platform, developed by the same group and designed to be used by the scientific and medical community in research projects, and into the Cancer Genome Interpreter, also developed by this group and which is more focused on clinical decision-making by medical oncologists.
BoostDM currently works with the mutational profiles of 28,000 genomes analyzed from 66 types of cancer. The scope of BoostDM will grow as a result of the foreseeable increase in publicly accessible cancer genomes.
An advance founded on evolutionary biology
To identify the mutations involved in cancer, the scientists based themselves on a key concept in evolution, namely positive selection. Mutations that drive the growth and development of cancer are found in higher numbers in distinct samples, compared to those that would occur randomly.
“We started from the premise that we only get to observe some mutations because the tumor cells with this mutation guide the development of the tumor, and we questioned what distinguishes these mutations from other possible mutations,” says Dr. Ferran Muiños, postdoctoral researcher and co-first author of the work. “Doing this analysis manually would be excessively laborious, but there are computational strategies that allow it to be organized systematically and efficiently,” he adds.
From the data, the proposed method learns what attributes are distinctive of the mutations that favor the development of cancer and this information is useful for the development of new therapeutic approaches.
https://youtube.com/watch?v=1Nq_rm_yudk%3Fcolor%3Dwhite
A computational model for each gene and type of cancer
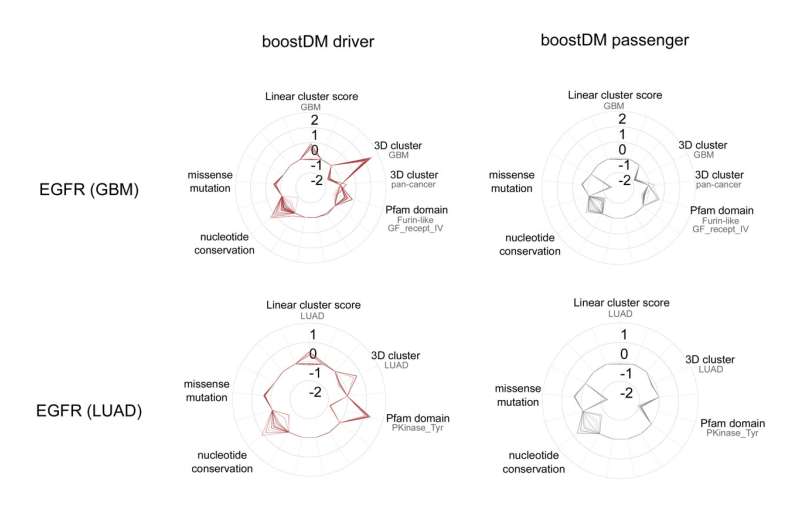
The tool that the researchers have developed has already generated 185 models to identify mutations in a specific gene in a given type of cancer. For example, it has produced a model that has identified all the possible mutations in the EGFR gene that trigger tumor development in some lung cancers, another model for the same gene in cases of glioblastoma (a type of cancer that affects the brain), etc.
As sequencing data on tumors become publicly accessible, it can be incorporated into the system, allowing it to generate new models for all cancer genes in the coming years.
Source: Read Full Article
