allegra dйfinition
Most severe acute respiratory syndrome coronavirus (SARS-CoV-2) mutations cause only mild harm. However, a small percentage of mutations can boost viral virulence and promote host-virus interactions that are critical to viral entry and infection. For example, SARS-CoV-2 spike protein mutations can significantly affect viral behavior, as the spike protein mediates viral attachment to host cell surface receptors.
It is crucial to monitor and reduce virus circulation to prevent the emergence of new SARS-CoV-2 variants that may not be sensitive to currently available vaccines and drugs. Despite widespread efforts that have been made by many countries around the world, mass vaccination campaigns have not achieved the required population coverage to prevent SARS-CoV-2 transmission.
 Study: How many relevant SARS-CoV-2 variants might we expect in the future? Image Credit: Naeblys / Shutterstock.com
Study: How many relevant SARS-CoV-2 variants might we expect in the future? Image Credit: Naeblys / Shutterstock.com
Classification of SARS-CoV-2 variants
In order to effectively control the pandemic, it is imperative to investigate the emergence and spread of variants and their impact on disease transmission and human health. The World Health Organization (WHO) has classified SARS-CoV-2 variants with a possible risk to public health into three distinct categories including variants under monitoring (VUMs), drug interaction nolvadex variants of interest (VOIs), and variants of concern (VOCs).
VUMs are variants with genetic mutations that alter viral characteristics, although the phenotypic or epidemiological impact of these mutations is not clear. VOIs have mutations that may affect infectivity, disease course, and diagnostic or therapeutic escape, which could lead to community transmission and a risk to global public health. VOCs are associated with increased transmissibility, virulence or disease severity, as well as the ability to decrease the effectiveness of interventions, diagnostics, therapeutics, and vaccines.
These variants may need to be reclassified over time as the virus undergoes continuous evolution. Quantifying the number of variants that may pose a potential risk for public health is crucial for future planning in the fight against viral epidemics.
About the study
In a recent study published on the preprint server medRxiv*, researchers fit data on the most relevant SARS-CoV-2 variants according to the WHO by exploiting a function that exclusively depends on the global number of infected cases since the start of the pandemic. Their fit permits fairly accurate estimation of the number of relevant SARS-CoV-2 variants that can emerge for a particular number of infected individuals around the world. This new approach can also predict the number of new relevant variants per ten million cases in any epidemiological situation.
The team gathered information on SARS-CoV-2 variants including variant characteristics reported by WHO, Phylogenetic Assignment of Named Global Outbreak (PANGO), and WHO classification, current relevance (VOC, VOI, or VUM), date and country of the first detection, the total number of global cases at the end of the month of detection, and a cumulative number of variants. PANGO is a nomenclature system used for naming and tracking genetic lineages of SARS-CoV-2. The numerical fit of this WHO data was obtained using the function v(N) = k x N⁄log N, where k is the constant of the numerical fit and is equal to 3.35 x 10−6.
“Our method depends critically on the WHO efficiency in tracking the most relevant SARS-CoV-2 variants.”
Study findings
The results of the study showed that the number of relevant variants of SARS-CoV-2, until November 2021, amounted to nearly 44. Comparatively, the number of new relevant variants per ten million cases was 1.64 in November 2021, which was a reduction of 28.4% from 2.29 that was reported in March 2020.
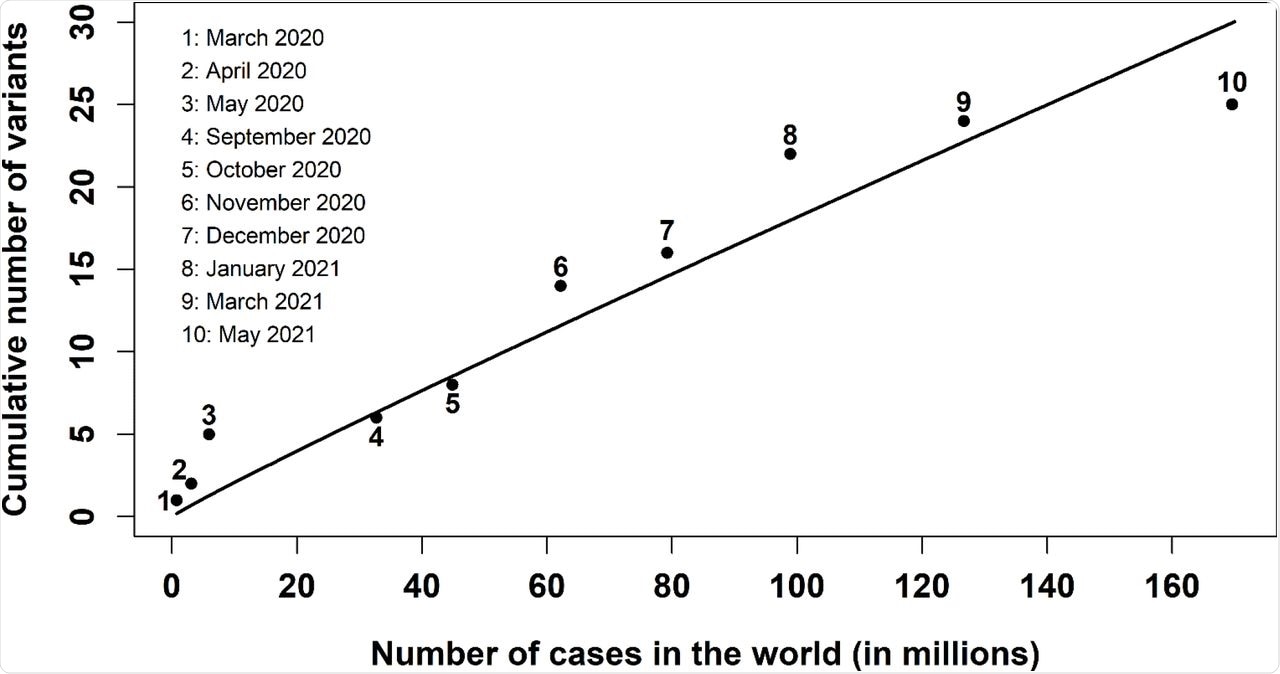 Cumulative number of relevant SARS-CoV-2 variants versus the cumulative number of cases in the world. The dots from 1 to 10 indicate the data reported by WHO [1, 2] from March 2020 to May 2021; the solid line represents the numerical fit υ = k · N/log N obtained with Wolfram Mathematica.
Cumulative number of relevant SARS-CoV-2 variants versus the cumulative number of cases in the world. The dots from 1 to 10 indicate the data reported by WHO [1, 2] from March 2020 to May 2021; the solid line represents the numerical fit υ = k · N/log N obtained with Wolfram Mathematica.
Until November 2021, the cumulative number of global COVID-19 cases was about 252 million, which corresponds to 43.7 relevant variants. This reflects almost 19 variants more than what was reported by WHO in May 2021.
Conclusions
The observations in this study revealed that the number of new relevant variants per ten million cases decreased very slowly with an increase in the cumulative number of cases. Thus, as long as the virus remains in circulation, new relevant SARS-CoV-2 variants will continue to emerge.
The authors built a mathematical model to calculate the number of relevant SARS-CoV-2 variants using the cumulative global number of infected cases. This model only focused on the relationship between the number of replications of the virus and the emergence of relevant variants and ignored all other factors involved in the diffusion of new variants.
The ability to predict the number of possible new relevant SARS-CoV-2 variants is very crucial in the future for optimal planning of vaccination campaigns, as new variants can alter viral characteristics and greatly affect the global management of the pandemic.
*Important notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Littera, R., & Melis, M. (2021). How many relevant SARS-CoV-2 variants might we expect in the future? medRixv. doi:10.1101/2021.11.17.21266463. https://www.medrxiv.org/content/10.1101/2021.11.17.21266463v1
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: Cell, Coronavirus, Coronavirus Disease COVID-19, Diagnostic, Diagnostics, Drugs, Evolution, Genetic, Pandemic, Protein, Public Health, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Therapeutics, Virus

Written by
Susha Cheriyedath
Susha has a Bachelor of Science (B.Sc.) degree in Chemistry and Master of Science (M.Sc) degree in Biochemistry from the University of Calicut, India. She always had a keen interest in medical and health science. As part of her masters degree, she specialized in Biochemistry, with an emphasis on Microbiology, Physiology, Biotechnology, and Nutrition. In her spare time, she loves to cook up a storm in the kitchen with her super-messy baking experiments.
Source: Read Full Article
