SARS-CoV-2 variants of concern exhibit divergent interactions with host proteases
In a recent study posted to the bioRxiv* preprint server, researchers at Weill Cornell Medical College assessed the cleavability of the spike (S) protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs) and related coronaviruses by several proteases of the host.
Host proteases cleave SARS-CoV-2 (and other coronaviruses) S at the boundary of S subunits 1 and 2 (S1/S2), thereby exposing peptides for the fusion of SARS-CoV-2 and host cell membranes which facilitates SARS-CoV-2 entry into the host. In a previous study, the authors demonstrated that host proteases, such as coagulation factors, cleave SARS-CoV-2 S and promote host invasion by SARS-CoV-2.
 Study: Evolution of host protease interactions among SARS-CoV-2 variants of concern and related coronaviruses. Image Credit NIAID
Study: Evolution of host protease interactions among SARS-CoV-2 variants of concern and related coronaviruses. Image Credit NIAID
About the study
In the present study, researchers extended their previous analysis by assessing the S1/S2 cleavage of SARS-CoV-2 and other coronaviruses by host proteases such as furin, transmembrane protease serine 2 (TMPRSS2), thrombin, and coagulation factor Xa. They used fluorescence resonance energy transfer (FRET)-based assays to explore the impact of SARS-CoV-2 VOC mutations on the corresponding S protein cleavage by proteases of the host. In addition, the cleavage of other coronaviruses’ S proteins by the proteases was also assessed.
Subsamples of databases such as Nextstrain and the global initiative on sharing all influenza data (GISAID) were examined for S codon 681 sequence divergence among SARS-CoV-2 VOCs. In particular, the team investigated the impact of mutations in SARS-CoV-2 variants on S1/S2 cleavage by host proteases by comparing the enzyme kinetics [maximum initial velocity (Vmax)] on substrates with P681 and P681H mutations corresponding to the Wuhan-Hu-1 (ancestral) strain and the Alpha VOC, respectively.
Furthermore, the activity of nafamostat in inhibiting proteases involved in SARS-CoV-2 entry was assessed. The effect of phosphorylation on host protease-mediated S1/S2 cleavability at serine (Ser) residues located in the vicinity of the S1/S2 cleavage site was determined using phosphorylated substrates corresponding to the S689, S680, and S686, residues.
In addition, the impact of post-translational modifications on S1/S2 cleavability was evaluated. Lastly, the evolution of protease-mediated S1/S2 cleavability across other related coronaviruses such as the SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), common cold virus (HCoV-OC43), the infectious bronchitis virus (IBV)-Beaudette, and the bat coronavirus (RatG13) was assessed.
Results
The S codon 681, which resides in the S1/S2 cleavage site, was of the highest entropy sites in the SARS-CoV-2 genome in the samples. The mutations P681H and P681R define the Alpha and Delta VOCs, respectively. While the P681H mutation enhanced the activity of factor Xa, there were no changes in the cleavage activity of furin, thrombin, and TMPRSS2.
However, the P681R mutation enhanced the Vmax values of the coagulation factor Xa-mediated SARS-CoV-2 S1/S2 cleavage and that of furin-mediated S1/S2 cleavage by 65% and 99%, respectively, in comparison to the Wuhan-Hu-1 strain. In contrast, thrombin and TMPRSS2 demonstrated reduced activity for the substrate expressing P681R.
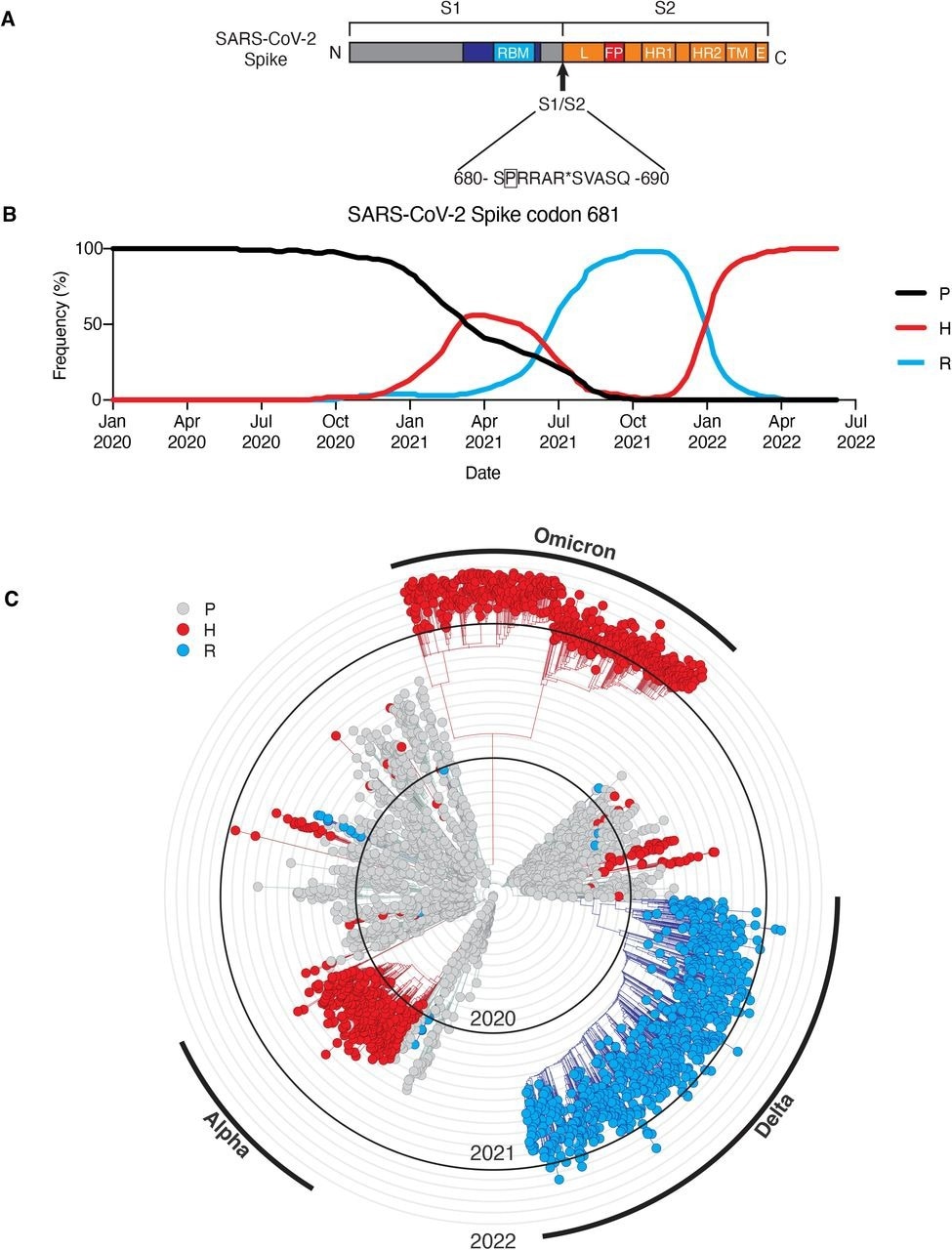
Sequence divergence of SARS-CoV-2 spike-681 among variants of concern. (A) Schematic of SARS-CoV-2 spike protein, highlighting position 681 adjacent to the S1/S2 site. Modified from (Kastenhuber et al., 2022). A subsampled collection of 3043 samples from between Dec 2109 and May 2022 from GISAID was obtained and visualized using Nextstrain (https://nextstrain.org/ncov) (Elbe and Buckland-Merrett, 2017; Hadfield et al., 2018). (B) Frequency of viral genomes sequenced with proline (black), histidine (red), or arginine (blue) at spike codon 681 by date of sample collection. (C) Phylogenic tree rendered by Nextstrain. Genotype at S681 of each sample is indicated by proline (gray), histidine (red), or arginine (blue). Branches corresponding to dominant variants of concern are highlighted in the outer ring.
Nafamostat inhibited coagulation factors, TMPRSS2, and other transmembrane serine proteases involved in SARS-CoV-2 entry. While factor Xa exhibited elevated Vmax for the variant substrates expressing the P681Rv and P681H and mutations, factor Xa demonstrated equivalent nafamostat sensitivity for cleaving the P681Rv and P681H substrates. Ser 680 phosphorylation at the P6 position located upstream of S1/S2 cleavage site, entirely terminated furin-mediated cleavage and moderately impacted (30% to 50% inhibition) S1/S cleave by factor Xa, thrombin, and TMPRSS2.
Ser 686 phosphorylation in the P1 position located near the S1/S2 cleavage site, strongly inhibited the activity of all proteases tested. Contrastingly, Ser 689 phosphorylation at the C-terminal P4 position impacted S1/S2 cleavability such that TMPRSS2 and factor Xa were inhibited moderately whereas thrombin was inhibited strongly, and on the contrary, increased furin-mediated cleavage was observed.
Post-translational modifications by phosphorylation significantly affected S1/S2 cleavability. Protease susceptibility was different for all the coronaviruses tested and only the cleavage site of SARS-CoV-2 demonstrated cleavage by the four proteases tested. Factor Xa demonstrated marked cleavability for the S1/S2 cleavage sites of SARS-CoV-2 and HCoV-OC43.
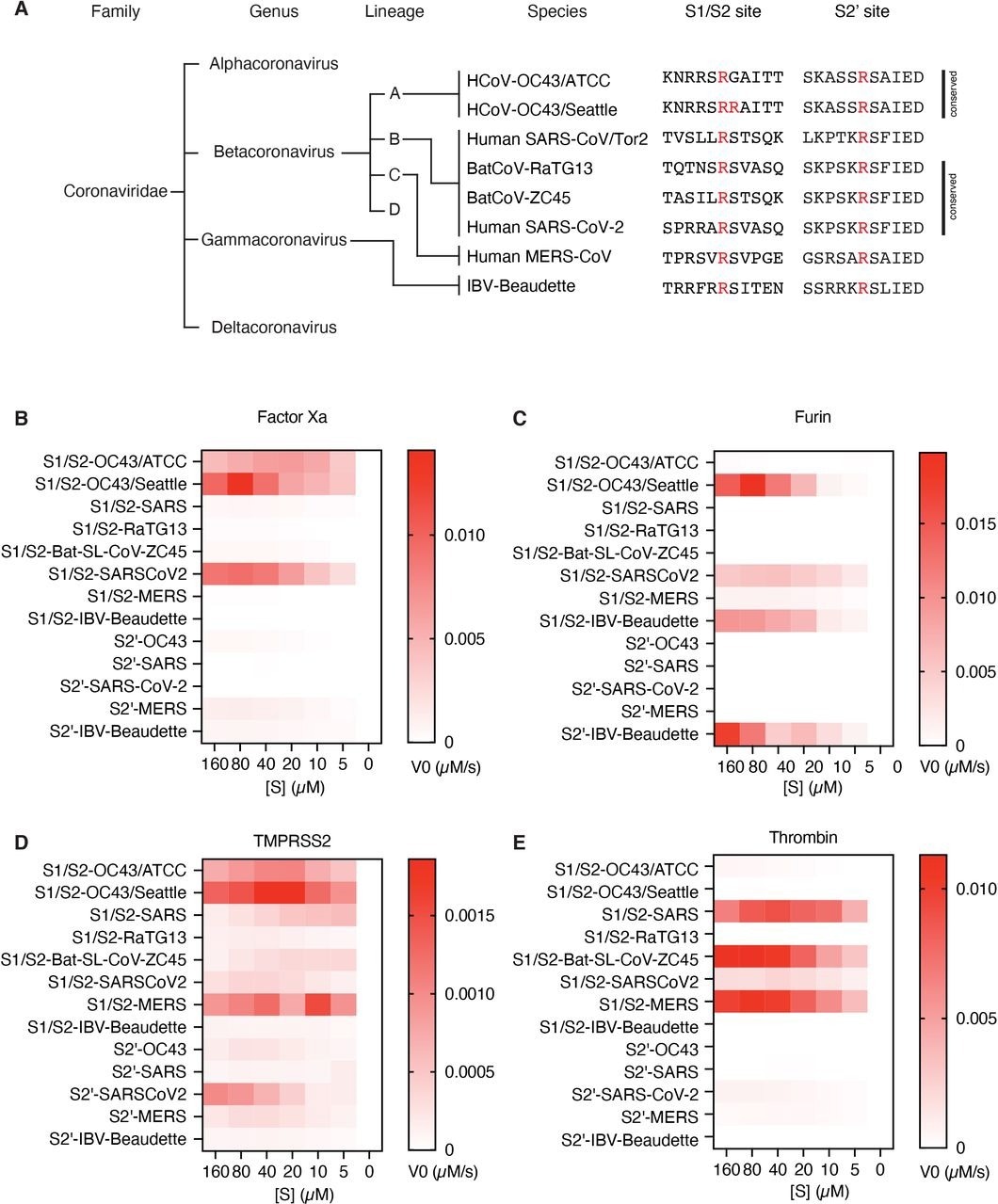
Proteolytic fingerprint of diverse coronavirus lineages. (A) Phylogenic relationship of a panel of coronaviruses with the corresponding aligned S1/S2 and S2’ cleavage sites. Heatmaps depicting the initial velocity V0 of cleavage of the indicated peptide substrates (rows) and concentrations (columns) by (B) factor Xa, (C) furin, (D) TMPRSS2, and (E) thrombin.
Furin showed efficacy against the HCoV-OC43 S1/S2-OC43/Seattle strain, but not against the S1/S2-OC43/ATCC strain of HCoV-OC43. In addition, furin efficiently cleaved the S1/S2 boundary and the S2 cleavage site of the IBV-Beaudette strain. Thrombin demonstrated cleavage of S proteins of MERS, SARS, and SARS-CoV-2, but did not substantially cleave RatG13.
TMPRSS2 cleaved the S proteins (at the S1/S2 and S2 cleavage sites) of several coronaviruses, although the cleavage efficacy was relatively low. In addition, the team observed convergence of distantly related coronaviruses to achieve common host protease interactions, including coagulation factors. This indicates that the use of host proteases has probably been a selection pressure through the evolution of coronaviruses.
Overall, the study findings showed that while SARS-CoV-2 VOCs exhibited divergent interactions between their S1/S2 cleavage sites and host proteases, convergent evolution of host protease-mediated cleavability was observed among different coronaviruses.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Evolution of host protease interactions among SARS-CoV-2 variants of concern and related coronaviruses. Edward R. Kastenhuber, Jared L. Johnson, Tomer M. Yaron, Marisa Mercadante, and Lewis C. Cantley. bioRxiv preprint 2022, DOI: https://doi.org/10.1101/2022.06.16.496428, https://www.biorxiv.org/content/10.1101/2022.06.16.496428v1
Posted in: Medical Research News | Disease/Infection News
Tags: Arginine, Bronchitis, Cell, Codon, Cold, Common Cold, Coronavirus, Coronavirus Disease COVID-19, Efficacy, Enzyme, Evolution, Fluorescence, Frequency, FRET, Genome, Histidine, Influenza, MERS-CoV, Mutation, Peptides, Phosphorylation, Proline, Protein, Respiratory, SARS, SARS-CoV-2, Serine, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Virus

Written by
Pooja Toshniwal Paharia
Dr. based clinical-radiological diagnosis and management of oral lesions and conditions and associated maxillofacial disorders.
Source: Read Full Article
